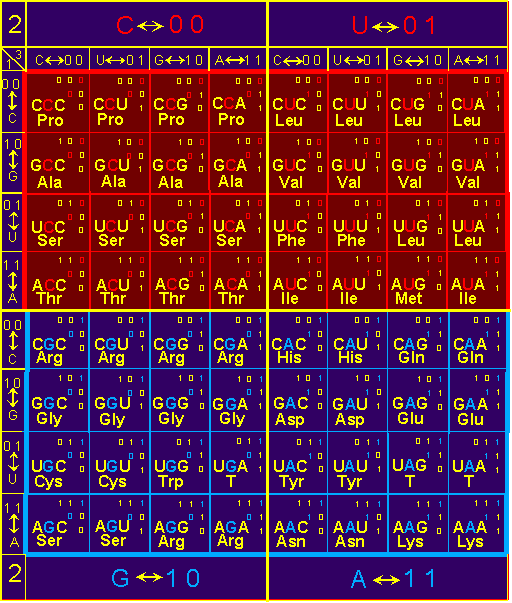
4.3. Assignment of amino acids side chains to blocks of a genetic code triplets
It's
time to try to answer the question posed at the end of Section 2.3. Why the amino acids are assigned exactly
to the given triplets, instead of others?
Let's lead comparison of the side chains,
classified on the basis of concepts of physical operators of connectivity and
anti- connectivity, with a table of the triplet genetic code (Fig. 38) obtained
by transformation of triangular matrixes of the
Supermatrix to triplets (Section 3.3.).
|
According to the definition given in Section 4.1.,
the operators of anti-connectivity – are
the side chains of amino acids of the protein, preventing the formation of hydrogen bonds NiH....Oi-4=C. Its properties should be non-polar amino acids. In the matrixes
describing the acyclic conformations of the graph and
protein, х3 = 0
(in pairs of variables x3x4: blocks 00 <---> C, 01
<---> U). As we have mentioned (Section 4.2.), side chains of amino acids that function as anti-connectivity operators belong in a genetic code to blocks of triplets C and U.
To see this, take a look at Figure
38. In block
C, as follows from the figure, are located
Pro (proline), Ala (alanine) - non-polar
amino acids not
capable to formation of hydrogen
bonds, Ser (serine), Thr (threonine) - weakly
polar amino acids that form
hydrogen bonds with the Oi-3=C. Similarly, in the block U are located
Leu (leucine), Val (valine), Phe (phenylalanine) and Leu (leucine), Ile (isoleucine) and Met (methionine) - non-polar amino acids not capable to formation of hydrogen bonds. |
|
|
According to Section 4.1., operators of connectivity
- the side chains of amino acids that contribute to fixing the hydrogen bond NiH....Oi-4=C. These
side chains should be able to form hydrogen bonds. In a matrix х3 = 1 (in
pairs of variables x3x4: blocks
10 <---> G, 11 <---> A), i.e.
these operators recreate cyclic 4-link protein fragments. The side chains of amino acids working as operators of connectivity (Section 4.2.), are in a genetic code in blocks of triplets G and A. In these blocks, triplets with G and A in the second position
encode polar amino acids, which
can form hydrogen bonds with the
Oi-4=C. Really (Fig.
38), in block G are
Arg (arginine), Gly (glycine), Cys (cysteine) and Trp (tryptophan) and Ser (serine) and Arg (arginine). The
block A contains the amino acids
His (histidine) and Gln (glutamine), Asp (aspartic) and Glu (glutamic acid), Tyr (tyrosine), Asn (asparagine) and Lys (lysine). |
|
|
Thus, there is full compliance of the group
properties of amino acids, considered as operators of anti-connectedness and connectedness, to those
conformations of proteins that are encoded by triplets in blocks of
the given structure of the genetic code. |
Fig. 38. The
table of triplet genetic code The triplets
received by coding of triangular matrixes of the Supermatrix, and amino acids
corresponding to these triplets are shown. |
Thus we have in general found the answer to the question at the beginning of this section: why the amino acids
correspond exactly to the given triplets, instead of others.
The answer is that the amino acids correspond to the
triplets, which encode the protein conformation, reconstituted by these amino acids in
the role of physical operators.
When considering the amino acid side chains in
Section 4.2.
you've probably noticed that as the length of the
side-chain increases hydrogen bond angle is changed: at the short side chains it is
less than 90 degrees, at the long
- more than 90 degrees. This could mean
that the side chains of different lengths may create
a "tug" in NiH...Oi-4=C, aimed
in different directions. This
observation was the starting point for further analysis of this sphere. Part of this analysis,
associated with the
reconstruction of symmetrical conformations of four-link graph and protein, carried
out in Section 4.4.