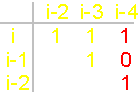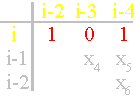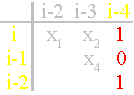4.4. Recreation
of symmetric conformations
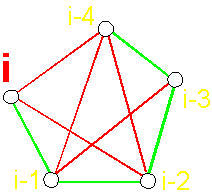
Earlier (see Section 3.2.) we saw, that in Supermatrix blocks by sides of the main diagonal are settled down symmetric conformation of the graph. For example, in the block
11 two symmetric graphs (Figure 39) are described by symmetric matrices 101111 и
111101:
|
|
|
|
|
|
Fig.
39. Symmetric conformation
of the 4-arc graph of the block
11 (a, c) and their
matrix descriptions (b, d). |
|||
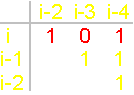
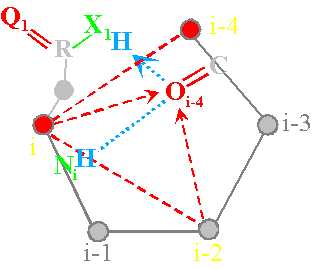
The question arises: what should be the operators recreating the symmetric conformation? Let’s consider the two operators of connectivity - with a short (left) and longer (right)
chain (Fig. 40):
|
|
|
|
|
|
Fig.
40. Operators of connectivity (a,
c), recreating the symmetric conformation of the protein and their matrix
description (b, d). |
|||
As shown in Figure 40,a,
the operator with a short chain at the expense of X1H forms a hydrogen bond with the Oi-4=C and contributes to the formation of a
hydrogen bond of main chain цепипNiH...Oi-4=C (atoms
i and i-4 are connected, in the matrix x3=1,
Fig. 40, b). It pulls the whole system up and to the left. Arising power lines are directed away from the atoms i and i-2.
Between them there is an edge of connectivity (in a
matrix x1=1). Therefore in a matrix is 1 0 1.
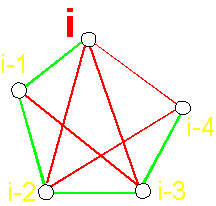
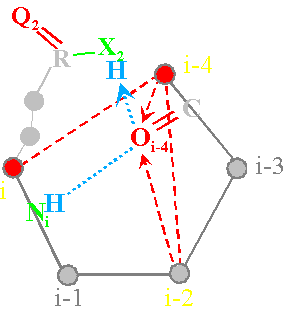
The operator with a longer chain (Fig.
40, c) forms a hydrogen bond with
the с Oi-4=C at the expense of X2H and also contributes to the
formation of hydrogen bonds the
main chain of NiH...Oi-4=C (atoms i and i-4 are
connected, x3=1). It
pulls system upwards and to the right. Power lines will be directed from atoms
i-2 and i-4 and between them there is the edge of connectivity (in a matrix x6=1, Fig. 40, d). In the third column of a matrix will
be 1 0 1.
Similar conclusions can be made by consideration of
operators of anti-connectivity. From our analysis follows, that symmetric conformation should be recreated by different physical
operators.
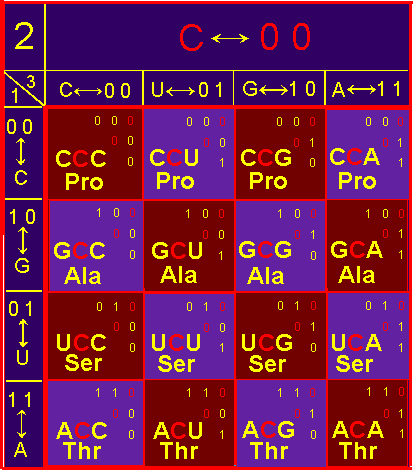
Let’s verify our conclusions in our table of the triplet genetic code.
|
|
|
|
Fig.
41. Analysis of the amino acids recreating symmetrical conformation in block С (a) and U (b). |
|
|
As shown in Figure 41, a, in the block С, symmetrically about the main
diagonal, going from top to bottom
from left to right, are located:
Ala - Pro, Ser - Pro, Thr - Pro, Ser - Ala, Thr - Ala, Thr - Ser. In pairs, all amino
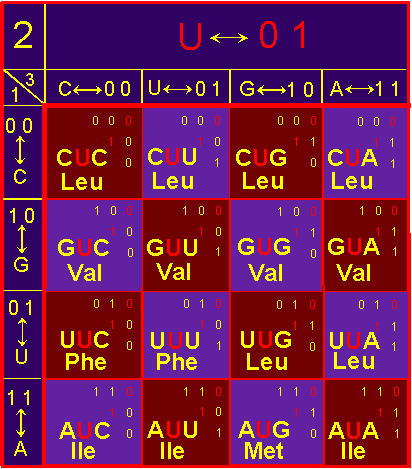
acids are different. In block U (fig. 41, b) symmetric conformation recreate
the following pairs: Val - Leu, Phe - Leu, Ile
- Leu, Phe - Val, Ile - Val, Met - Leu. All amino acids of pairs are
different.
|
||||||
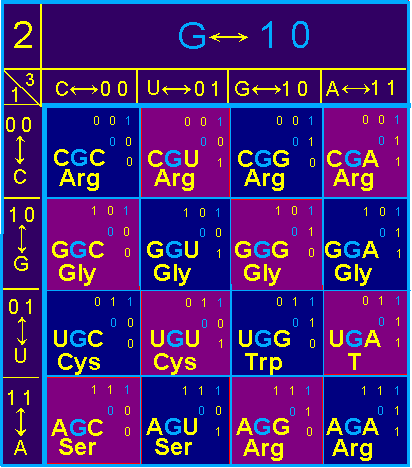
In block G
(Fig. 42, a) symmetric position relative to the main diagonal is occupied: Gly - Arg, Cys - Arg, Ser - Arg, Cys - Gly, Ser -
Gly, Arg - T. Amino acids in pairs are different. T is a stop signal.
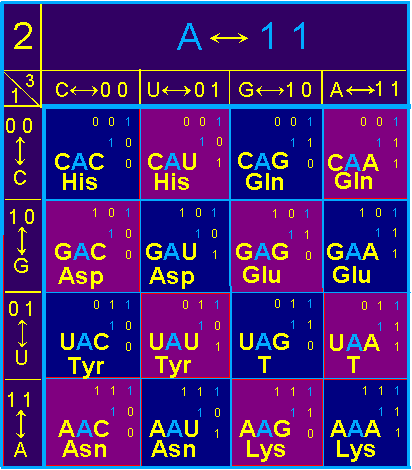
In block
A symmetric conformations are recreated by pairs:
Asp - His, Tyr - Gln, Asn - Gln, Tyr - Glu, Asn - Glu, Lys - T. Amino acids of pairs are different.
So,
in a real code the arrangement
of amino acids is so, that in symmetric pairs there are different side chains. And this despite the fact that in the block U
there are 6 leucine, and in block G - 6 arginine..
Thus, our conclusions that the symmetrical conformation of the protein must be recreated by different physical operators do not contradict the
properties of the triplet genetic code.
In the course of the
analysis it has been revealed, that various side chains (e.g., identical in properties,
but different in length amino acids
asparagine and glutamine) can have different directions of the applied force (in other words,
different vectors of action). This
was the starting point to a special
analysis of the physical
operators in the area of the main chain bond цепипNiH...Oi-4=C and resulted
in the development of model of molecular vector
machine of proteins. On the basis of these representations the highly
effective way of predicting of secondary structure of proteins, which has become a patent
basis, is developed. More in detail it is possible to familiarise with these
questions on our site «Molecular vector machine of proteins»: http://vector-machine.narod.ru/
We wish you successful scientific traveling!