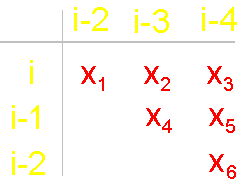3.1. Definition of initial concepts
In articles [6,15-20], and also in the monograph [8]
we developed the theory of topological coding of chain polymers.
According to these works, we
name the chain polymers not branched out polymers consisting of repeating units. Proteins belong to the class of chain polymers, therefore
this theory has been easily adapted directly to the proteins [18].
3.1.1. Concept of a link
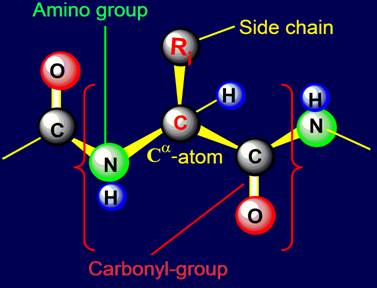
Usually the link of protein called
amino acid residue (side chain)
associated with the alpha-carbon atom to
which are attached on one side
- an amino group, on the other - a carbonyl group (Figure 18,a).
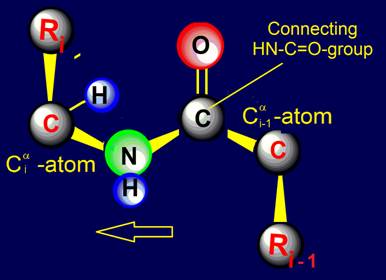
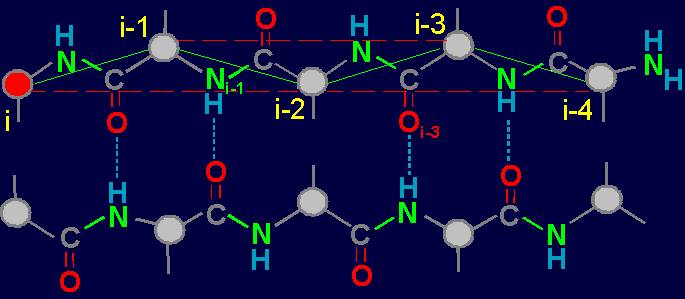
Within the framework of our approach a link is the fragment of the protein consisting of
two alpha-carbon atoms
with attached to them side chains Ri, Ri-1, and group HN–C=O connecting them (fig. 18, b).
|
|
|
|
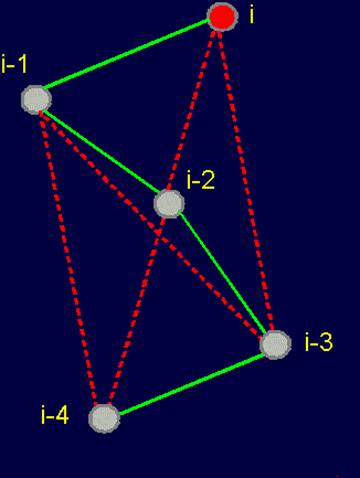
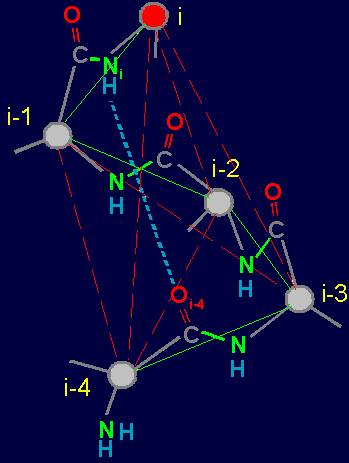
Fig. 18. To
definition of concept of a link. a - the accepted concept of
amino acid link in the protein; b -
concept of a link within the
framework of the
theory of topological coding of proteins. |
|
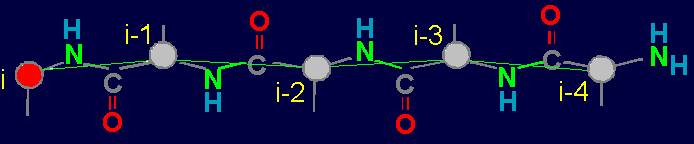
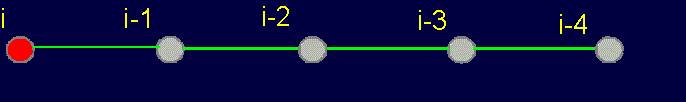
3.1.2. Four-arc chain graph
–analogue of protein pentafragment
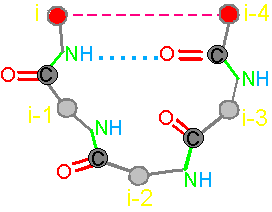
In the framework of the stated theory the fragment of five amino acids has been allocated
as an elementary object for theoretical analysis
(Fig. 19, a). We
call it a pentafragment or a 4-link fragment of the
protein. As seen in Fig. 19, a,
this fragment contains 5 alpha-carbon
atoms (hence - pentafragment), which are linked by four connecting groups
HN–C=O, which corresponds to four links within the limits of our concepts (Section 3.1.1.).
The
choice of pentafragment for analysis is
caused by the
fact that it is minimal and the most common fragment of the protein, which can
form a hydrogen bond between the NiH-group belonging to the i-th
alpha-carbon atom and the carbonyl group O i-4=Ci-4 related to i-4-th alpha-carbon
atom. Protein fragments by hydrogen bond can fix its conformation in
a cycle. We call this property a connectivity.
|
|
|
|
||
|
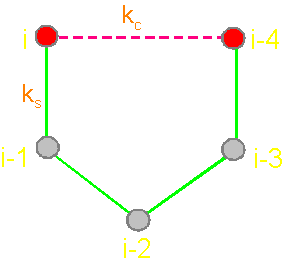
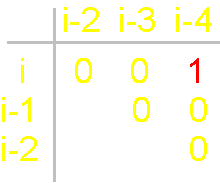
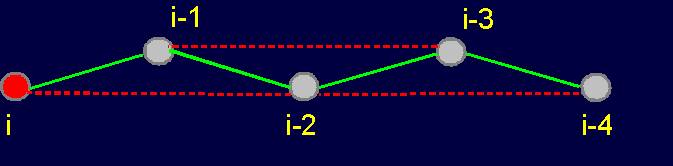
Fig.
19. Basic concepts
of the theory of topological coding
of proteins. a - 4-link fragment of the protein (pentafragment); a dotted line denotes the fixation of i-th and i-4-th atoms in the formation of hydrogen bonds in the cycle; b -
analogue of a protein pentafragment - 4-arc chain graph. Solid lines - structural
edges, dashed line - edge of connectivity; c
- matrix describing the conformation of the above polymer fragments
and the graph, d - a general view of the matrix. |
||||
|
Explanation of designations in Figures 19, a – 19, d. |
||||
|
Fig. 19, a. |
Fig. 19, b |
Fig. 19, c, d |
||
|
In
this figure, the alpha-carbon
atoms of amino acids (circles with rim) are numbered as i, i–1, i–2, i–3, i–4. Peptide bonds – HN–C=O-groups. The link includes: two alpha-carbon atoms and the peptide bond connecting them (e.g., i – i–1). The length of the protein link – a relatively constant value. This
fragment is fixed by a hydrogen bond formed by two peptide bonds: O=C–NH.....O=C–NH The atoms i and i-4 lose their mobility and
are connected. |
The solid lines connecting the vertices of the
graph correspond to protein
link connecting the neighboring alpha-carbon atoms. Let’s
call them the structural edges. The link of the
graph consists of two adjacent vertices and structural edge between
them and is analogue of a protein link. The length of the structural edge is constant and is characterized
by a constant ks. For the description of connected vertices,
we introduce the concept of "edge of connectivity", which connects non-adjacent
vertices. Some of such edges
may be identical length
and some - have a set
of characteristic lengths, designated kс. In our illustration
the edge of connectivity (dotted
line) connecting the vertices i
and i-4. |
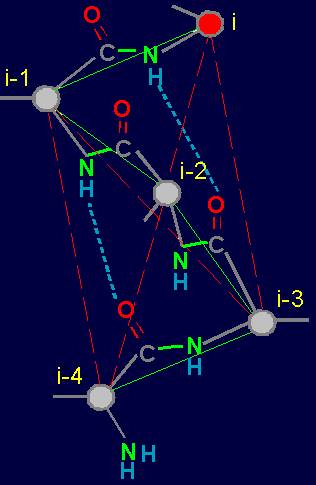
To
construct the matrix we write vertices from left: i, i-1, i-2, and at
the top - vertices i-2, i-3, i-4. On crossing
of lines and the columns going from the vertices, in the matrix will
be written: 1 - if there is a connectivity edge,
and 0 - if it
is not. There are only 6 such crossings in the
matrix: i - i-2, i -
i-3, i - i-4, i-1 - i-3,
i-1 - i-4, i-2 - i-4. In
our example (Fig. 19, b) the edge
of connectivity bind vertices i - i-4, therefore in a matrix on crossing of i-th
row and i-4th column is 1. Other edges of connectivity in the graph are not present, owing to what in other crossings
there are zeros. The general form of the matrices of the
six elements is shown on fig. 19,
d. x1 - x6 are
the variables capable to accept values 0 and
1 (Boolean variables). Sometimes will be used also record in
one line: x1x2x3x4x5x6. |
||
Using the mathematical analogue - 4-arc
chain graph allows
us to consider all its possible conformations.
However, before we go to them, it is necessary to remind the basic conformations, which can accept the protein chain,
and how these can be described with the
help of graphs and matrices.
3.1.3. Typical structures of proteins, their graphs and matrix description
Four types of conformations the most commonly found in proteins [21, 22]:
- Weakly
connected conformation;
- A
stretched conformation in the
form of beta-structure;
- Strained helical conformation 310, in which the hydrogen bond is formed between atoms NiH … Oi-3=C of the two peptide groups;
- Alpha-helical
conformation, energetically the most
favorable, in which the hydrogen bond
is formed between the NiH … Oi-4=C.
Below are pentafragments of these
conformations, the four-arc fragments of
their graphs and descriptions
in the form of upper triangular
matrices. Pay attention to the matrices:
we see them in the blocks of Supermatrix, which will be presented in
the following section.
|
|
|
|
|
|
|
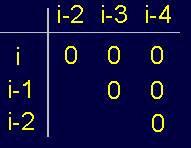
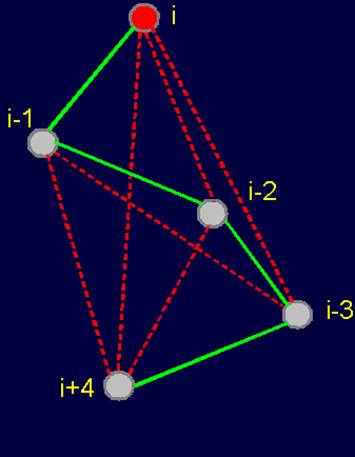
Fig.
20. Pentafragment of weakly-connected structure of the protein
(a), its 4-arc
graph (b) and matrix
description (c). |
|
A
fragment of a weakly- connected protein structure (Fig. 20,a) is represented by a 4-arc graph, which has no edges of connectivity [ (Fig. 20,b), so describing this conformation matrix contains only zeros (Fig. 20,c).
|
|
|
|
|
|
|
Fig.
21. Beta-structural protein pentafragment (a), its 4-arc
graph (b) and matrix
description (c). |
|
In the
layered beta-structure of the protein (Fig. 21, a) fixation of
non-adjacent alpha-carbon atoms is
due to hydrogen bonds. For
example, N i-1H-group of i-1-th amino acid and C=Oi-3-group
of i-3-th form hydrogen
bonds with the neighboring chain.
As a result, the alpha-carbon atoms of i-1- i-3-rd amino acids are connected
(shown in dotted lines in Fig.
21, a). In a 4-arc
graph of this structure (Fig. 21, b) the edges of connectivity join
vertices i - i-2, i-1 - i-3 and i-2
- i-4, which is reflected in the matrix (Fig. 21, c ): values of
1 in the corresponding crossings of rows and columns.
|
|
|
|
|
Fig.
22. Pentafragment of protein conformation in
the helix 310 (a), its 4-arc graph (b)
and matrix description (c). |
||
In
the helix 310 two hydrogen-bond system of the HN–C=O-groups are formed, which fix its
conformation (Fig. 22,a). They provide
a connection of almost all the alpha-carbon atoms of 4-link fragment,
with the exception of i-- i-4-th,
which is clearly visible on the graph (Fig.
22,b) . In the matrix (Fig. 22,c)
for this pair of vertices is 0.
|
|
|
|
|
Fig.
23. Alpha-helical protein pentafragment (a), its 4-arc
graph (b) and matrix
description (s). |
||
The typical structure of the protein is an alpha-helix, 4-link fragment of which is shown in Figure 23 as well. There is only one
hydrogen bond between NiH
and Oi-4=C, fixing the atoms i-th
and i-4-th. However, in more extended fragment the other two HN–C=O-groups are also involved in the fixation of the alpha-carbon atoms. As a consequence,
all the atoms of alpha-helical fragments are connected,
as shown in the graph (Fig. 23, b). All
variables in the matrix are set
to 1 (Fig. 23, c).
The following questions arise:
- How much
of connected conformations of 4-link protein fragments, their graphs and matrix of 6 elements
can exist?
- Can
they be to classified and
arranged in a table any way?
In order to obtain answers to these questions we conducted
this work on a 4-arc
graphs, and have
constructed supermatrix of its conformations and that of triangular matrices describing these the
conformations (section
3.2.) .